7.2 Model evaluation(not part of the official work)
This section is just further exploration, I barely know what I am doing here. –Hung Nguyen
The exploration focuses on the model3 with all of the variables.
P1 <- ggplot(data=data.frame(model3$residuals), aes(y=model3$residuals, x=model3$fitted.values)) +
geom_point(alpha = 0.1) +
ggtitle("Residual Plot") +
xlab("Predicted Values") +
ylab("Residuals") +
geom_hline(yintercept = mean(model3$residuals), color = "red")+
geom_smooth(method = "loess") + #Locally Weighted Least Squares Regression
annotate("text",
x = 0.9, y = 0.5,
label = paste("Cov:",round(cov(model3$residuals,model3$fitted.values), digits =2)),
size = 4, color = "purple",
)
P2 <- ggplot(data=data.frame(model3$residuals), aes(x=model3$residuals)) +
geom_histogram() +
ggtitle("Histogram of Residuals") +
xlab("Residual")
P3 <- ggplot(data=data.frame(model3$residuals), aes(sample = model3$residuals)) +
stat_qq(alpha = 0.1) +
stat_qq_line(color = "red") +
xlab("Normal Quantiles") +
ylab("Residual Quantiles") +
ggtitle("QQ Plot")
grid.arrange(arrangeGrob(P1, P2, ncol =2), arrangeGrob(P3,textGrob(label = "Zero mean assumption is true, which is no suprise. \nThere is Heteroskedasticity in the residual. Residual \ndistribution have a slight left tail, but not too extreme.\nThe blue curve in the first plot raise doubt about \n the Zero Conditional Mean Assumption.", just = "left", x = unit(0.1, "npc"), y = unit(0.5, "npc"), gp=gpar(fontsize=10)) ,ncol =2))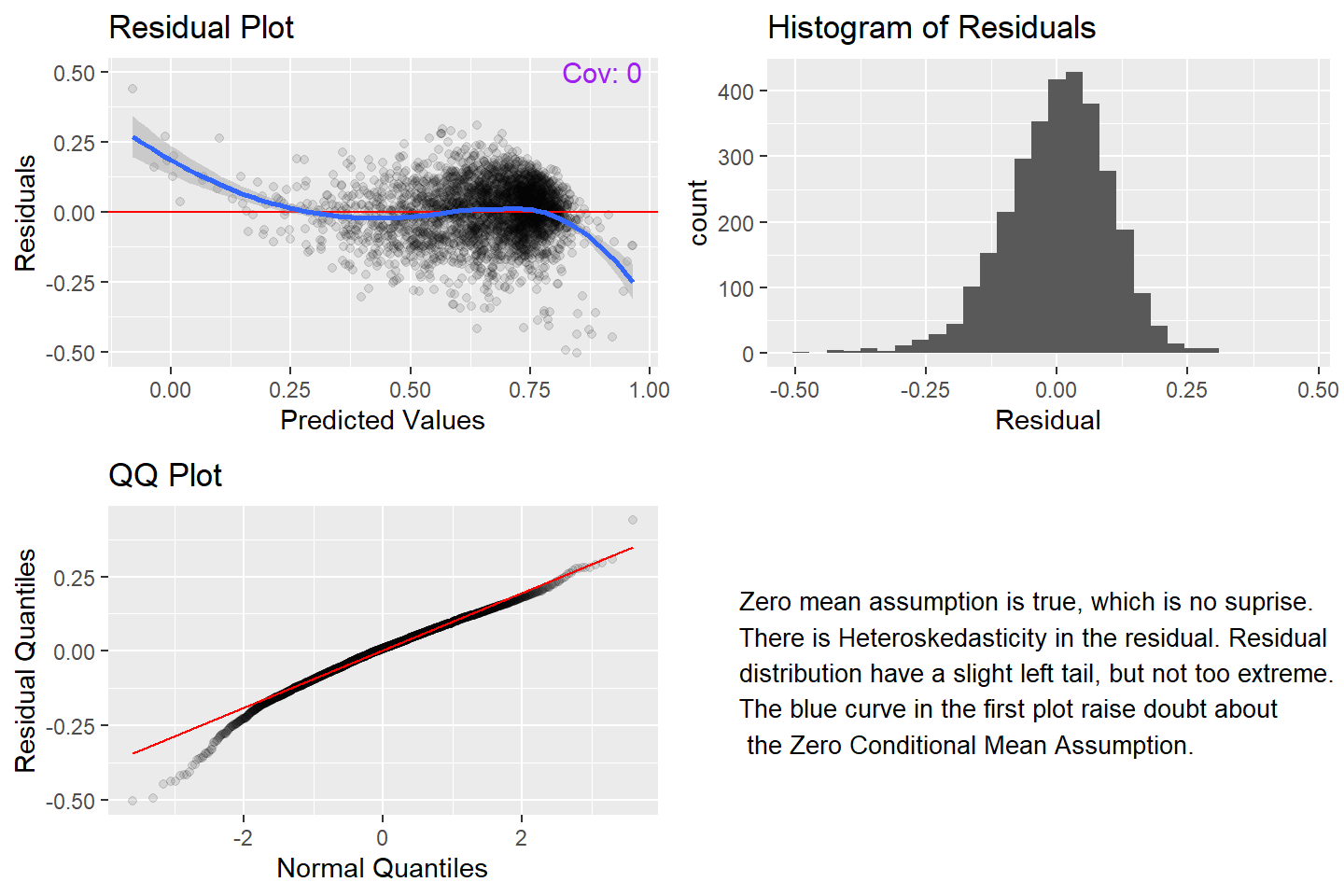
Next we observe the relation between the residual and each of the variables.
v1 <- ggplot(data=data.frame(model3$residuals), aes(y=model3$residuals, x=model3$model$median_rent)) +
geom_point(alpha = 0.1) +
geom_smooth(method = "loess") +
labs(x="Median Rent", y= "Residuals") +
annotate("text",
x = 2125, y = -0.5,
label = paste("Cov:",round(cov(model3$residuals,model3$model$median_rent), digits =2)),
size = 4, color = "purple",
)
v2 <- ggplot(data=data.frame(model3$residuals), aes(y=model3$residuals, x=model3$model$median_age)) +
geom_point(alpha = 0.1) +
geom_smooth(method = "loess") +
labs(x="Median Age", y= "Residuals") +
annotate("text",
x = 65, y = -0.5,
label = paste("Cov:",round(cov(model3$residuals,model3$model$median_age), digits =2)),
size = 4, color = "purple",
)
v3 <- ggplot(data=data.frame(model3$residuals), aes(y=model3$residuals, x=model3$model$median_income)) +
geom_point(alpha = 0.1) +
geom_smooth(method = "loess") +
labs(x="Median Income", y= "Residuals") +
annotate("text",
x = 62500, y = -0.5,
label = paste("Cov:",round(cov(model3$residuals,model3$model$median_income), digits =2)),
size = 4, color = "purple",
)
v4 <- ggplot(data=data.frame(model3$residuals), aes(y=model3$residuals, x=model3$model$perc_hs)) +
geom_point(alpha = 0.1) +
geom_smooth(method = "loess") +
labs(x="People without High School(%)", y= "Residuals") +
annotate("text",
x = 60, y = 0.30,
label = paste("Cov:",round(cov(model3$residuals,model3$model$perc_hs), digits =2)),
size = 4, color = "purple",
)
v5 <- ggplot(data=data.frame(model3$residuals), aes(y=model3$residuals, x=model3$model$perc_rent)) +
geom_point(alpha = 0.1) +
geom_smooth(method = "loess") +
labs(x="People renting(%)", y= "Residuals") +
annotate("text",
x = 30, y = -0.5,
label = paste("Cov:",round(cov(model3$residuals,model3$model$perc_rent), digits =2)),
size = 4, color = "purple",
)
v6 <- ggplot(data=data.frame(model3$residuals), aes(y=model3$residuals, x=model3$model$perc_white)) +
geom_point(alpha = 0.1) +
geom_smooth(method = "loess") +
labs(x="White population(%)", y= "Residuals") +
annotate("text",
x = 10, y = -0.5,
label = paste("Cov:",round(cov(model3$residuals,model3$model$perc_white), digits =2)),
size = 4, color = "purple",
)
v7 <- ggplot(data=data.frame(model3$residuals), aes(y=model3$residuals, x=model3$model$perc_below_pov)) +
geom_point(alpha = 0.1) +
geom_smooth(method = "loess") +
labs(x="Population below poverty line(%)", y= "Residuals") +
annotate("text",
x = 50, y = 0.3,
label = paste("Cov:",round(cov(model3$residuals,model3$model$perc_below_pov), digits =2)),
size = 4, color = "purple",
)
v8 <- ggplot(data=data.frame(model3$residuals), aes(y=model3$residuals, x=model3$model$perc_doc)) +
geom_point(alpha = 0.1) +
geom_smooth(method = "loess") +
labs(x="Population with doctorate(%)", y= "Residuals") +
annotate("text",
x = 15, y = -0.35,
label = paste("Cov:",round(cov(model3$residuals,model3$model$perc_doc), digits =2)),
size = 4, color = "purple",
)
grid.arrange(arrangeGrob(v1,v2,v3,v4,v5,v6,v7,v8, ncol = 2), top ="Residual Plot over Variables")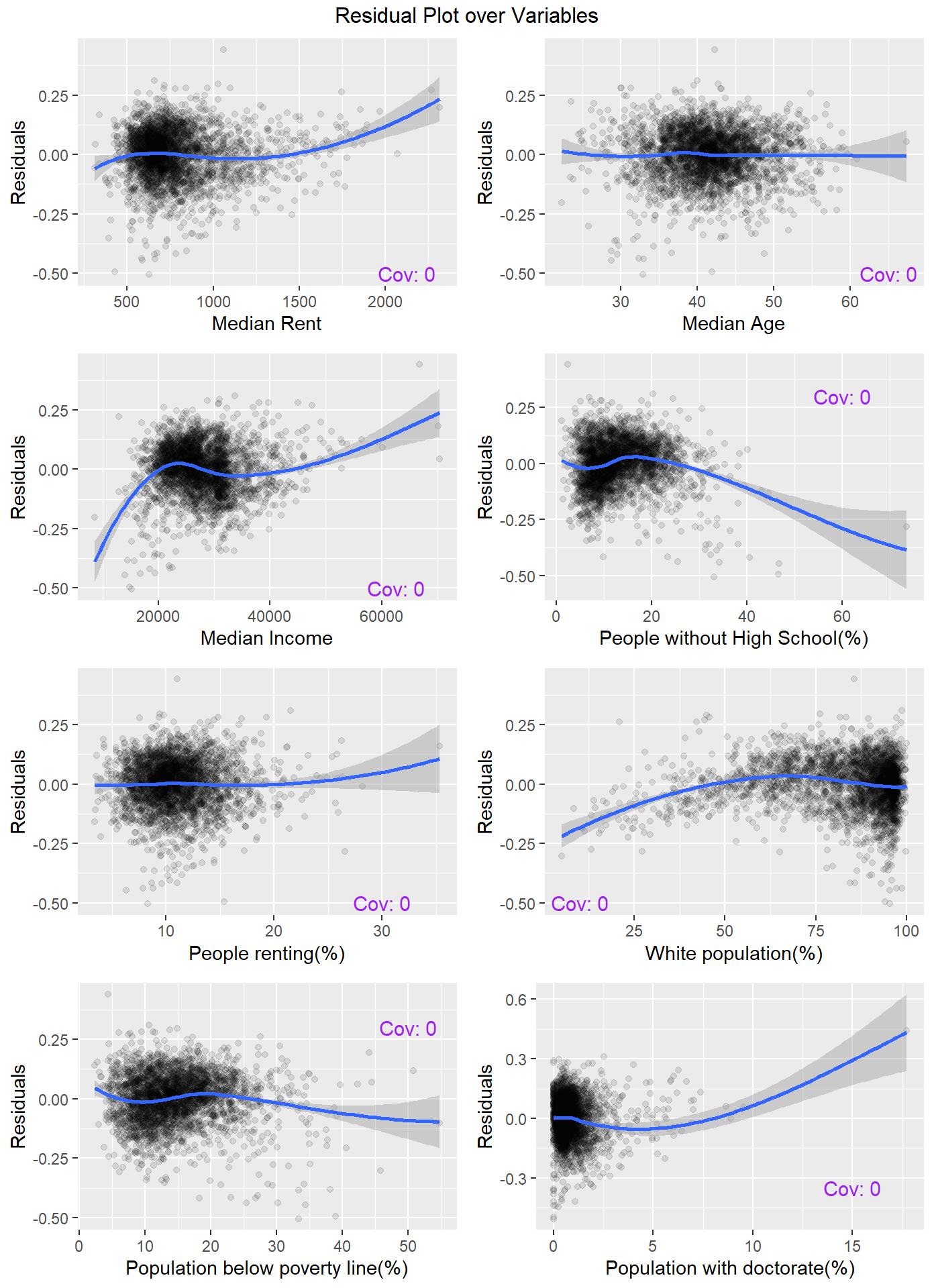
The covariances being zero is just a property of the OLS estimator.